|
OF CELLS AND CYCLOTRONS
One of the principal objects of theoretical research in any department
of knowledge is to find the point of view from which the subject appears in its greatest simplicity.
J. Willard Gibbs
The editor of Nature, the late John Maddox, tried many times through his editorials to warn molecular biology
that failing to 'get physical' will only serve to enable the 'hi-jack of DNA' by the physicists and, as signature
to that, it is then a physicist, Dr John Male, who was tasked by default with looking into electromagnetic
fields and biological systems effects, who reports finding that:
' ... the particular genes activated and the
kinds of proteins synthesized in the cells appeared to depend on the frequency and waveform of the magnetic field applied
... An intriguing feature of some of these effects is that they seem to require the presence of a static magnetic field (comparable
with the geomagnetic field) modulated by a parallel alternating field of similar amplitude whose precise frequency is critical
... the observed values ... physicists will immediately recognise as the so called cyclotron frequency of the free
ion ... this is a remarkable finding ... However, it is difficult
to imagine that anything like this cyclotron resonance process could be going on inside a living cell'. (Male,
1992)
'Ion cyclotron resonance occurs when ions move in a steady magnetic field such as that of
the Earth. The field deflects them sideways and they go into orbit around its lines of force at a characteristic 'resonant'
frequency which depends on the charge/mass ratio of the ion and the strength of the steady field. Exposing them to an
oscillating electric or magnetic field at their resonant frequency lets them absorb its energy and they gradually increase
the size of their orbits and their energy of motion. The resonant frequency for potassium in the Earth's magnetic
field is close to 16 Hz.' (Goldsworthy, 2007).
Such findings - new to Dr Male, but long known in bioelectromagnetics - of course
beg the question and a variety of researches, most significantly including those of AR Liboff, have been
directed towards identifying the source of such resonance, mostly associated with either a variety of molecules
that might be capable within themselves of producing cyclotron resonance or with various membrane and calcium cascade
effects. No works have been identified to date which deal with the possibility of it being the genetic material, the chromosomes
themselves, as a possible source.
In order to best illustrate to a general readership - for
which this site must necessarily be intended - what this means in a more pragmatic sense is explored by simple
illustration of how we might identify a 'cyclotron' type source within the cell and we can best do this
by lashing together our own model of a cyclotron, a sort of rainy Sunday afternoon at the dining room table type modelling
...
Illustrated above are the required parts of a cyclotron as
gleaned from one of the science texts of early post-quantum physics, when things were still new and pictures of the first
machines of this emerging technology proudly offered! From these various parts we can piece together a cartoon
of the ideal cyclotron so as to obtain (working from top to bottom of the cartoon construction):
(i) a
proton or ion source
(ii) a means of channelling and storing same
(iii) a defined path
for their accelerated flight
(iv) a trapped target atom and
(v) an 'acceptor' or 'detector' system
In essence the principle of the machine is to fire accelerated and, therefore, high-energy particles at
a trapped atom and then use the gathered energetic output data of the 'collisions', 'kicks' or 'perturbations' to
better understand the particle/atom interaction in the given conditions.
Using this cartoon as some sort of analogue,
we have to ask if any such thing can be found in the genome? In order to explore this we first have to refer
to the biological data.
CHROMOSOME BASICS
Whilst
not wishing to oversimplify the case, there is a need for clarity that
is best served by
simple illustration. We start at the level of gross morphology
in terms of the chromosomal material as this is the level at which the gedanken
model is worked - and, of course, one level at which the system
itself must work. The
detailed structure of DNA will be dealt with as the need arises.
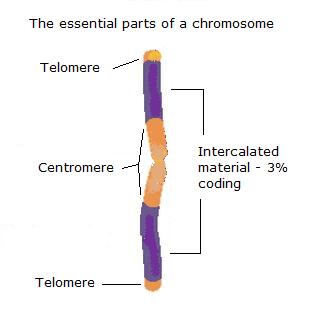
The
drawing above offers crude outline for the basic structure of
the chromosome, at least as far as need be understood for the model. The
essential structures are the two
telomeres and the centromere, the material that is then intercalated between these being more variable and, as we now know thanks to the human genome
mapping project, predominantly non-coding, with regions realising biological functionality then intercalated within that.
This material arrangement constitutes the
chromosome arm.
The illustration is an adaptation from web material for teaching and in this original, 'textbook' description
the orange coloured material is termed 'heterochromatin', 'where the DNA is more condensed and usually
there is not much transcriptional activity', i.e., was classically regarded as 'junk DNA', 'some of which
material remains condensed throughout the cell cycle'. The purple material or 'euchromatin' marks 'where the "active" genes are generally
found and usually this region is much less condensed'. It is, of course, grossly exaggerated
here. Some 97% of any arm is pretty much really 'junk DNA' and should therefore mostly be orange with a few
- approximately 25,000 - very thin purple lines scattered about. So 'classical' texts now need that update.
There is a third required component within this material array that in fact underpins DNA function and duplication
and that is the means of access to the bases and their sequence within the DNA helix for either mRNA production or DNA
replication. Either of these functions is a transient state within the larger dynamics of the nucleus and the cell
and we will return to them later.
MODELLING THE GENOME
|
|
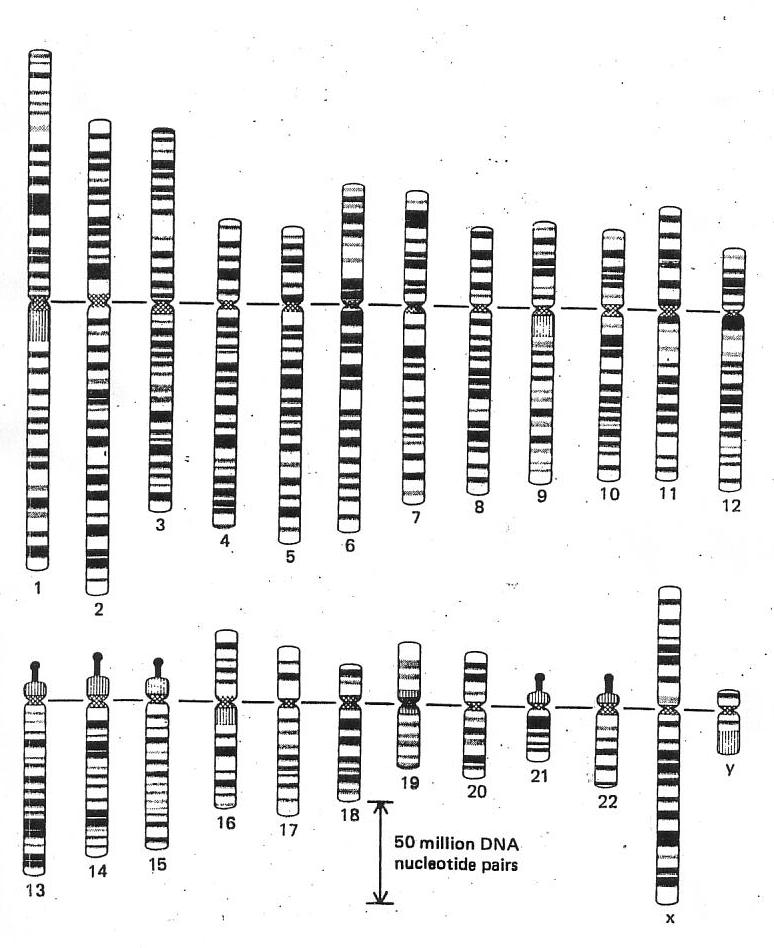
| From Alberts et al., 2nd edition, 1989 (and others) |
The gedanken experiment model that is to be developed here is a deconstruction
based on this graphic presentation of the human genome taken from Alberts et al., The Molecular
Biology of the Cell (2nd ed. and others). Derived from the earlier work of Ute Franke et al., it is a 'blocking-up'
of the original, fine-scale data.
Two immediate questions
arise:
- why is it laid out like this?
Whatever the 'scientific' reason offered, this ordering
obscures any 'natural', simple, pattern
that recognises it as a system accreting crystalline
mass over time. If we were to start with
a simple bacterial genome of 1m base pairs, then the human genome, with 3,000m base pairs,
can be simply understood as such an accretion system. It is only logical, therefore, to lay
the set out simply by crystal length.
- why does the
system spin down to 23 pairs of chromosomes?
This second question has to do with where QLS
naturally has to start which is to ask the seemingly
ludicrous question of 'How many types of atom are involved in living systems working?', i.e., what are we looking at in terms of componental complexity?
We could trawl all the
books and average our results or, instead, just take any reference to act as initiator for the model and this latter is the path taken here where one of the early texts in
'bioinorganic chemistry' (a developing discipline
through the late 80s) from Frausto da Silva & Williams (1991) declares interest to be focussed around
some 25 elements significantly involved in cellular
organisation. This list of 25 elements does not, of course, include Carbon which has its own extraordinary organic chemistry.
This then
suggests a model using 26 system elements. Life has trite, humorous or minimal description
as being a 'carbon-based system dependent
on water' - hydrogen and oxygen - which then gives a founding set of 3 elements as the 'system itself', if you like, leaving 23 elements needing ordination and control in
the complex environment of the cell.
This 1:1 mapping of 23
free elements and 23 chromosome pairs is very suggestive in terms of information theory, at least, but we know that it
is not, could never be - indeed imagine how much
more complex evolutionary study would be if
it were the case - that an organism could embed in a single
chromosome its total potassium functionality, for instance, but we know that
it is there, necessarily, somewhere, as a network distributed across the set.
Two further points
need be made here, the first having to do with the nature of the model as an abstraction of a graphic presentation.
The original itself of course has some level of error and the blocking-up of Alberts et al., introduces further error of course.
The model made here then offers a third level of error. We then need to ask whether the results that seem to spin out
quite naturally are 'artifact' or indicative of a robust system? How many errors can you make before you end
up at the truth anyway? There is a need to look at the inverse of Grover's quantum theory-based search algorithm
for unsorted databases to determine, instead of the shortest route to the desired end, the maximum route that might be taken
before the desired end is reached anyway. What happens also then when new steps are introduced to the route?
The other point of significance that needs to be made is that the fundamental components that are the system itself, i.e.,
carbon and water, are both functionally tetrahedral and, therefore, fractally 'conformal' or 'congruent'.
These founding three also have symmetric oxidation states - Carbon 8, Oxygen 4 and Hydrogen 2. The only other significant
atomic species with symmetric oxidation states is Sodium with 2.
RECONSTRUCTION
HARMONIC
The classical layout
of the genome as given above seems to have no natural logic beyond the science of the day and so we are free to explore
any more rational construction. Laying the set out by size seems a first and most reasonable step.
|
|
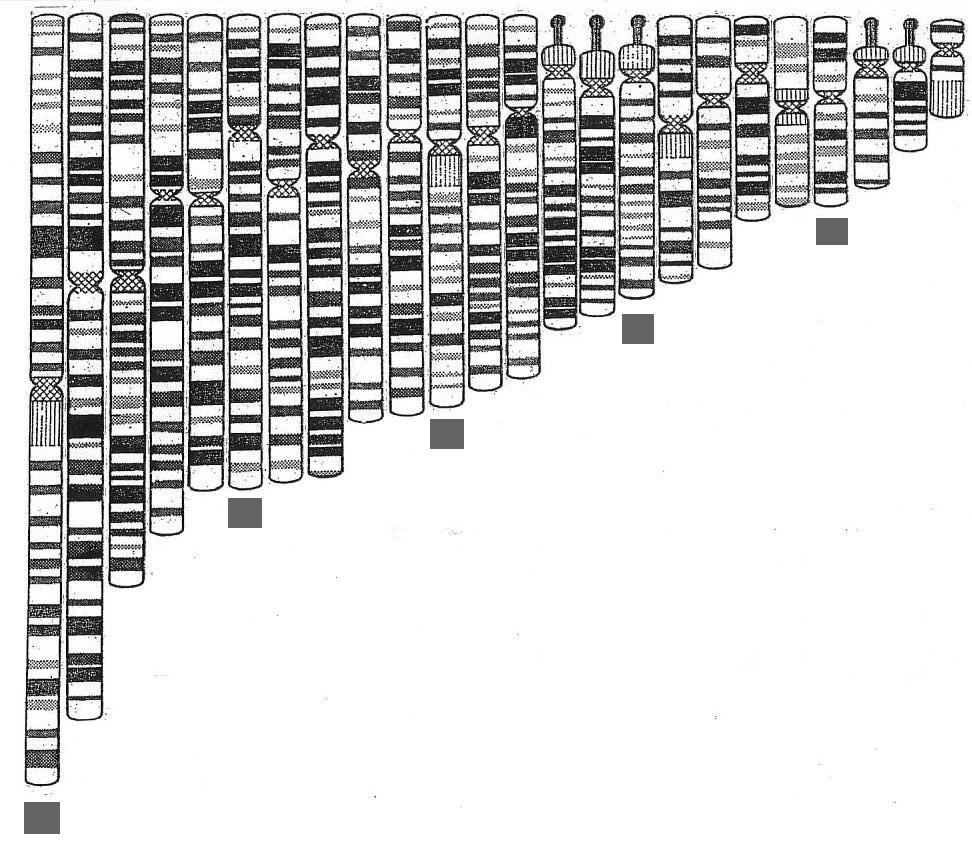
| The set laid out more naturally with members of one of five sub-sets indicated by the squares |
Laid out in natural order like this two things immediately become apparent: - the set has some harmonic ordering, like organ pipes laid out in logical array
before
installation
- this is not the fundamental set. There are five chromosomes
- 13, 14,
15, 21 and 22 - which carry the highly condensed arms that are multiple
repeats of sequences coding
for ribosomal RNA (rRNA) - and, as such,
represent highly periodic crystalline regions - and these stand apart, just
by their morphology, from the rest of the set indicating:
* The five condensed arms can be taken to indicate that we have 5 sub-systems operating
in parallel. This possibility is confirmed when we make up the implied 5 sub-sets by the simplest logical
means within the ordering by size of the group. If we take the five longest chromosomes of the set as the
longest of each of 5 sub-sets and then recruit subsequent set members by fives, it can be seen that each of
the sub-sets generated will in itself constitute an harmonic set - see
figure above. That such a simple logic is valid seems confirmed by the fact that it does result in the 5 highly
condensed arm chromosomes self assorting so that each is in a different sub-set - see figure below.
Further, there
is also circumstantial evidence for this 'five-foldedness' of the system in the fact that the average number
of signal protein binding sites in the upstream or downstream promoter regions of genes is five - which makes sense
in terms of information sharing in five parallel processing systems - each sub-system would inform itself and
the four other systems, each then having five-fold input and ensuring the synchrony of working across the sub-sets.
|
|
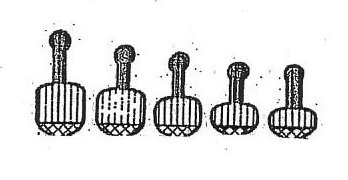
| The coloured dots are used to illustrate the self-assortment of the 5 condensed arm chromosomes |
* The second point of note is that simply by the
structure of these five chromosomes we might fairly propose that the single or individual arms in fact form the basic units
of the fundamental set.
|
|
| The human genome in single arm array shows an improved harmonic |
As has earlier been shown the double structure
chromosome set, i.e., the normal two armed structure, offers only a rather crude and lumpen harmonic pattern to
emerge while the single arm array, generated by separating the arms at the chromosome waist (above) makes clearer
the harmonic arrangement of the DNA crystals when in this condensed state characteristic of the cellular metaphase.
The five more highly condensed rRNA arms are also harmonically arranged, as illustrated below.
|
|
| The five highly condensed arms show their own harmonic ... |
Just this first step of reconstruction brings to
the fore the significance of the five 'condensed arm' or 'half-chromosomes' that seem to indicate five
sub-sets operating in parallel and it is these structures that immediately also identify themselves as possible analogues
for our cartoon cyclotron.
The above illustrates the general structural parallel, but
we need to take this cartoon analogue piece by piece to check out how far this extends.
> ANALOGUE WORKING
|